New Targets for Pesticide Discovery: Protein-Protein Interactions
By Jonathan Gressel, LEC Partners (formerly LEC Partners) & Weizmann Institute of Science and HiCap Formulations (Israel) Ltd.
Modern pesticides provide a healthy, safe food supply
The twentieth-century discovery of small organic molecule pesticides revolutionized agriculture and food production. Pesticides such as herbicides replaced manual and mechanical cultivation, while fungicides and insecticides severely lowered wastage due to insects and pathogens. These allowed the marketing of healthier, mycotoxin-free produce and superseded the previously used sulfuric acid, chlorate, arsenicals, and mercurials.

But lo, resistance evolved
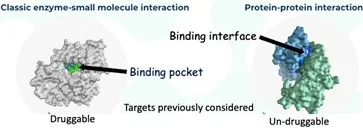
These small organic molecules were more specific, and this specificity led to the downfall of many pesticides; it allowed nature to evolve resistance mechanisms to overcome their effectiveness. The specificity is derived from the ability of these small molecules to bind into a deep binding pocket within a single enzyme. This, in turn, allowed a single mutation in their target to preclude pesticide binding, conferring resistance. Resistance and more stringent regulatory requirements resulted in the removal of many pesticides from the market, putting more evolutionary pressure on the evolution of resistance on the remaining compounds, which targeted fewer enzymes. As much as the agrochemical industry tried, they had diminishing returns, finding new enzymes targeting deep binding pockets. For over three decades, no new targets were discovered for herbicides.
Pharma had the same problem – devised new solutions
The pharmaceutical industry faced the same problem – resistance evolving to their drugs. They took on a new direction, targeting interactions between proteins where there are many new targets. These targets had been considered ‘undruggable’ in the past. Many proteins are made up of peptide sub-units, which are separately synthesized and combined in a highly specific manner using relatively weak binding forces. Some metabolic pathways work in cascades where the product of one enzyme is immediately metabolized by another enzyme, which must be in close contact, i.e., there must be a protein-protein interaction by weak binding forces. Unlike the deep binding pockets previously targeted, pharma had to find chemicals that would disrupt the more diffuse broad interface where the peptides or proteins bind. This required using the vast resources available to the pharmaceutical industry. They first had to crystallize the interacting proteins, then perform high-resolution X-ray crystallography and interpret the structures to find ‘hot spots’ where the interactions were strongest, not as mundane as it might seem from this short description. Pharma successfully found and commercialized short peptides and small organic molecule drugs that disrupt protein-protein interactions.
Ag-Chem start-up to the rescue
Could this approach be used to find new pesticides? Enter a group of enzyme and computational chemists, graduates of the pharmaceutical industry where they had worked on drug design targeting ‘undruggable’ protein-protein interactions. In 2019, they founded Projini AgChem to try their luck to find small molecules that will target ‘undruggable’ targets in pests. They chose to find weed killers, as herbicides account for more than half of the pesticide market, and it is easy to find target enzymes that do not occur in mammals, facilitating registration. They chose the two enzyme protein-protein interactions that produce the sulfur amino acid cysteine as a target. Cysteine is a key amino acid, providing the sulfur for all other organic sulfur compounds in all living organisms. After analyzing the interaction for hot spots, they developed computational tools to begin screening a virtual library of existing and yet-to-be-synthesized 40 million chemicals using the coordinates of the ‘hot spot.’ They first had 400,000 hits, then increasingly strengthened their criteria using an iterative process to get 250 hits eventually. Unlike what the agrochemical industry usually does, all this was without synthesizing a chemical or spraying a plant. Only then did they manually choose chemicals, which they tested on the interacting proteins they made in transgenic yeast. Some of these compounds kill plants and are being tested in the field.
These new inhibitors incalcitrant to the evolution of resistance
There is an advantage to targeting protein-protein interactions – it is harder for organisms to evolve resistance at the target. A single mutation that repels pesticide binding to one of the proteins will also repel its binding to the other protein, inactivating the enzyme. That is – unless the other protein has a complementary mutation. The frequency of two simultaneous complementary mutations is so low as to be insignificant. Is there a future for finding pesticides disrupting protein-protein interactions? Hindsight says yes – we now know that two herbicides found by random screening, paraquat and 2,4-D that have been in widespread use for over half a century act by disrupting protein-protein interactions. And guess what, despite resistance evolving to most other herbicides, no healthy target-site resistant plants have been found in the field to 2,4-D or paraquat. The time has come for the Ag-Chem industry to change its paradigms – there are a lot of new computational tools and targets to find new chemicals, requiring far less physical screening. The new chemicals found that target protein-protein interactions are likely to have a longer commercial life because they are calcitrant to the evolution of target site resistance.
About the Author
Jonathan Gressel received his M.Sc. and Ph.D. from the University of Wisconsin-Madison and has since been at the Weizmann Institute of Science, now as an emeritus professor. He also co-founded and was for three years Chief Scientist of TransAlgae, Ltd., a company devoted to transgenically domesticating microalgae as a source of high protein animal feed and biofuel, cultivated using seawater and industrial carbon dioxide. Dr. Gressel is involved in determining how plant sciences can contribute to the world bioeconomy, especially in food security, and has collaborated worldwide. He studied metabolic controls, especially by anti-metabolites and pesticides, and the evolution of resistance. He conceived and helped develop a system to control parasitic Striga (witchweed) using herbicide-treated crop seeds, which are commercialized throughout Africa. He has developed genetic engineering biosafety measures to mitigate the transgene movement. Due to his interest in turning basic research into applications, he has had long-term consultancies for multinational agrochemical producers as well as with Israeli ag-biotech companies, served on project evaluation panels for the NSF, EU, BARD, and has performed project evaluation for two VCs, authored >325 scientific papers and book chapters ,and is the author or editor of nine books dealing with these issues. His latest authored book is “Genetic Glass Ceilings – Transgenics for Crop Biodiversity” (Johns Hopkins Press). He has received an honorary fellowship in major professional societies based on his research and was awarded the 2010 “Israel Prize,” the highest accolade from the Israeli Government for his Agricultural Research.
About LEC Partners
LEC Partners was founded as LEC Partners in 1995 and has grown to become the world’s premier consulting group specializing in the bioeconomy, with over 180 experts worldwide. The group’s experts are renowned, hand-selected leaders, with over 97% holding advanced degrees and averaging over 30 years in their respective fields.
Have some questions?
Not sure where to start?
Let's start a conversation. We're here to help you navigate
the bioeconomy with confidence.